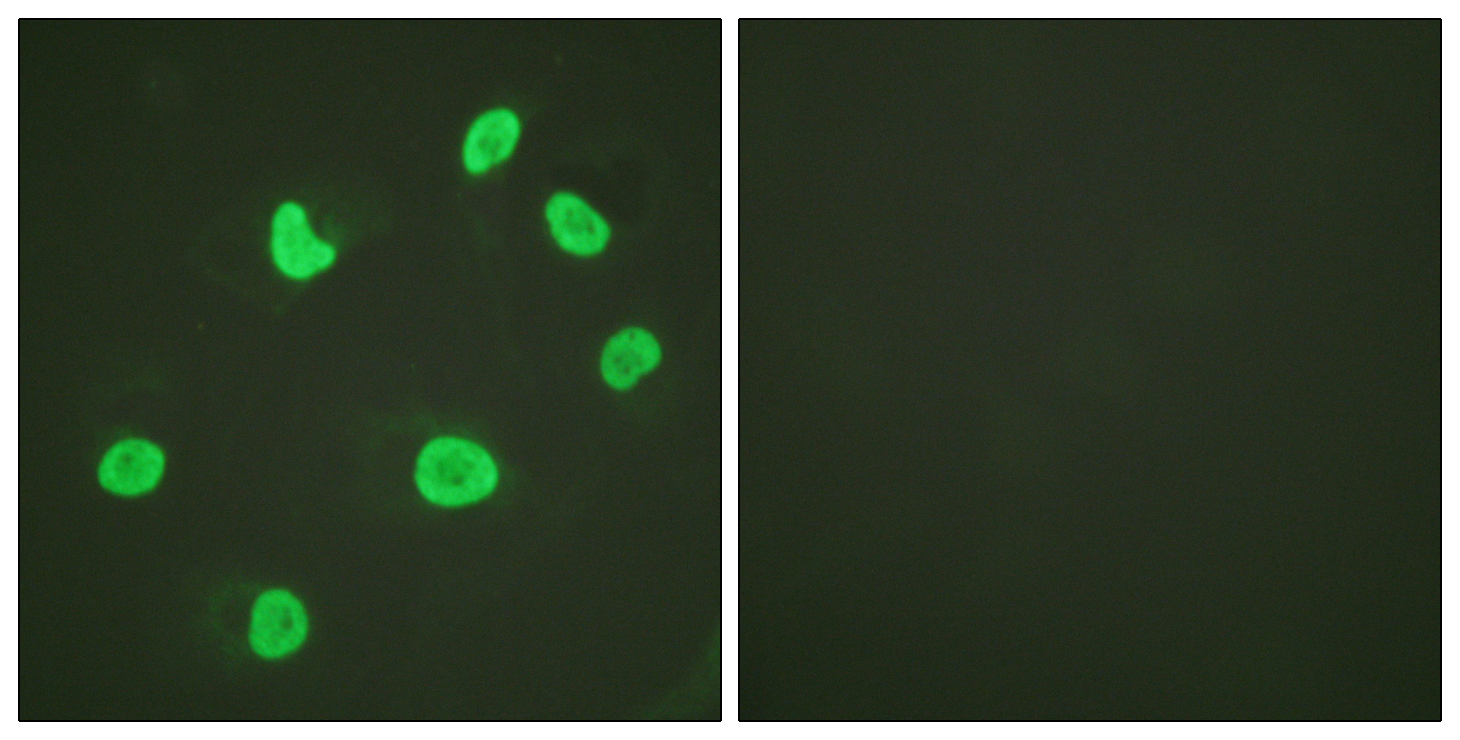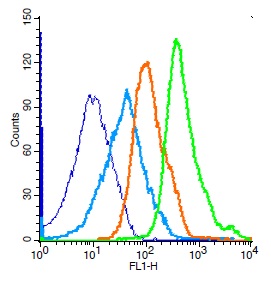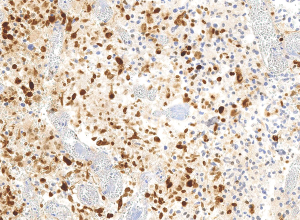
IHC of clinically validated FFPE Glioblastoma tumor tissues with H3.3 G34R expression, using anti-histone H3.3 G34R rabbit monoclonal antibody, Clone RM240. Clinically validated to be Positive for H3.3 G34R Mutation. Image courtesy of Dr. Sebastian Brandn
anti-Histone H3.3 G34R Mutant (human), Rabbit Monoclonal (RM240)
REV-31-1120-00
ApplicationsWestern Blot, ELISA, ImmunoHistoChemistry
Product group Antibodies
ReactivityHuman
TargetH3-3A
Overview
- SupplierRevMAb Biosciences
- Product Nameanti-Histone H3.3 G34R Mutant (human), Rabbit Monoclonal (RM240)
- Delivery Days Customer2
- ApplicationsWestern Blot, ELISA, ImmunoHistoChemistry
- CertificationResearch Use Only
- ClonalityMonoclonal
- Clone IDRM240
- Gene ID3020
- Target nameH3-3A
- Target descriptionH3.3 histone A
- Target synonymsBRYLIB1, H3.3A, H3F3, H3F3A, histone H3.3, H3 histone family member 3A, H3 histone, family 3A
- HostRabbit
- IsotypeIgG
- Protein IDP84243
- Protein NameHistone H3.3
- Scientific DescriptionHistone H3 is one of the DNA-binding proteins found in the chromatin of all eukaryotic cells. H3 along with four core histone proteins binds to DNA forming the structure of the nucleosome. Histones play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. Histone H3 has three main variants, H3.1 and H3.2, which are deposited in chromatin only during DNA replication and H3.3, which is replication independent and is found primarily in the regions of active transcription and heterochromatin. Post translationally, histones are modified in a variety of ways to either directly change the chromatin structure or allow for the binding of specific transcription factors. The N-terminal tail of histone H3 protrudes from the globular nucleosome core and can undergo several different types of post-translational modification that influence cellular processes. These modifications include the covalent attachment of methyl or acetyl groups to lysine and arginine amino acids and the phosphorylation of serine or threonine. Histone modifications are one form of epigenetic information that relate closely to gene regulation. Aberrant histone methylation caused by alteration in chromatin-modifying enzymes has long been implicated in cancers. Recently, recurrent histone mutations have been identified in multiple cancers and have been shown to impede histone methylation. All identified histone mutations (including H3K4M, H3K9M, H3K27M, H3K36M, and H3G34V/R/W) result in amino acid substitution at/near a lysine residue that is a target of methylation. - Recombinant Antibody. This antibody reacts to the Histone H3.3 G34R mutant. No cross reactivity with wild type Histone H3.3. Applications: ELISA, WB, IHC. Source: Rabbit. Liquid. 50% Glycerol/PBS with 1% BSA and 0.09% sodium azide. Histone H3 is one of the DNA-binding proteins found in the chromatin of all eukaryotic cells. H3 along with four core histone proteins binds to DNA forming the structure of the nucleosome. Histones play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. Histone H3 has three main variants, H3.1 and H3.2, which are deposited in chromatin only during DNA replication and H3.3, which is replication independent and is found primarily in the regions of active transcription and heterochromatin. Post translationally, histones are modified in a variety of ways to either directly change the chromatin structure or allow for the binding of specific transcription factors. The N-terminal tail of histone H3 protrudes from the globular nucleosome core and can undergo several different types of post-translational modification that influence cellular processes. These modifications include the covalent attachment of methyl or acetyl groups to lysine and arginine amino acids and the phosphorylation of serine or threonine. Histone modifications are one form of epigenetic information that relate closely to gene regulation. Aberrant histone methylation caused by alteration in chromatin-modifying enzymes has long been implicated in cancers. Recently, recurrent histone mutations have been identified in multiple cancers and have been shown to impede histone methylation. All identified histone mutations (including H3K4M, H3K9M, H3K27M, H3K36M, and H3G34V/R/W) result in amino acid substitution at/near a lysine residue that is a target of methylation.
- ReactivityHuman
- Storage Instruction-20°C,2°C to 8°C
- UNSPSC41116161