
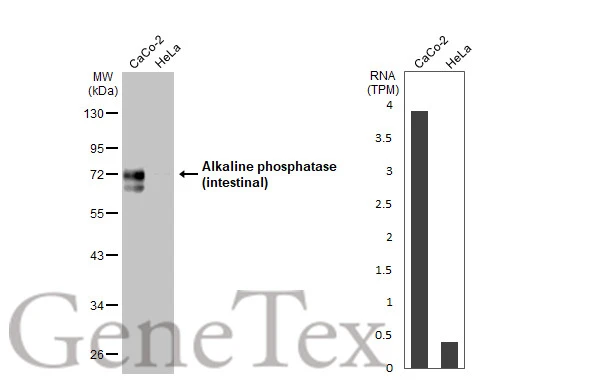
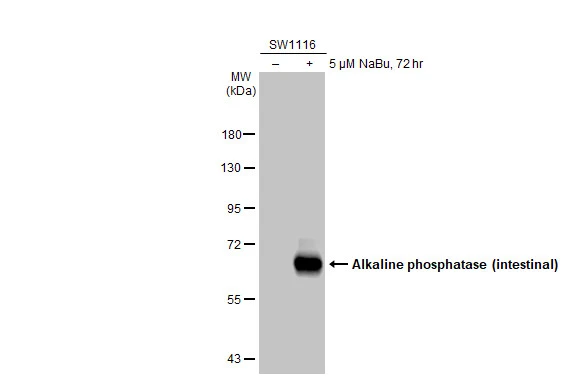
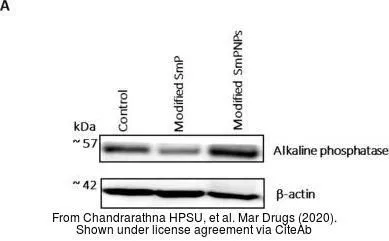
Non-transfected (–) and transfected (+) 293T whole cell extracts (30 μg) were separated by 10% SDS-PAGE, and the membrane was blotted with Alkaline phosphatase (intestinal) antibody (GTX112100) diluted at 1:5000. The HRP-conjugated anti-rabbit IgG antibody (GTX213110-01) was used to detect the primary antibody.
Alkaline phosphatase (intestinal) antibody
GTX112100
ApplicationsWestern Blot, ELISA, ImmunoHistoChemistry, ImmunoHistoChemistry Frozen, ImmunoHistoChemistry Paraffin
Product group Antibodies
ReactivityHuman, Mouse
TargetALPI
Overview
- SupplierGeneTex
- Product NameAlkaline phosphatase (intestinal) antibody
- Delivery Days Customer9
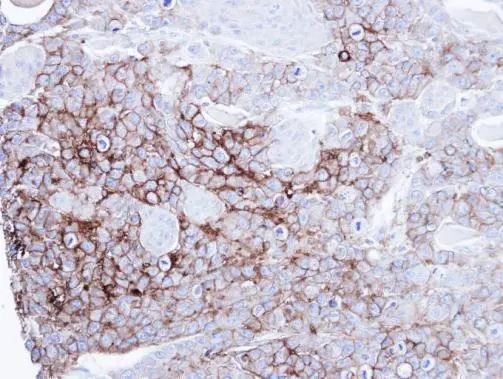
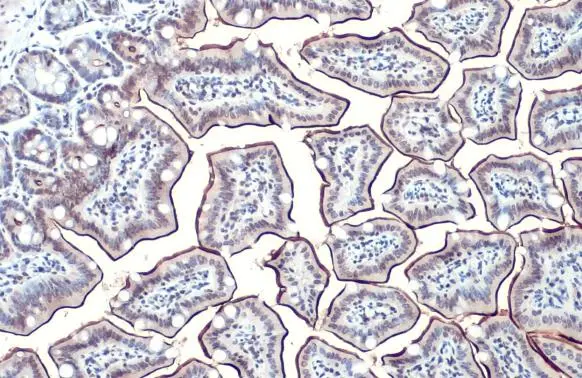
- Application Supplier NoteWB: 1:500-1:3000. IHC-P: 1:100-1:1000. *Optimal dilutions/concentrations should be determined by the researcher.Not tested in other applications.
- ApplicationsWestern Blot, ELISA, ImmunoHistoChemistry, ImmunoHistoChemistry Frozen, ImmunoHistoChemistry Paraffin
- CertificationResearch Use Only
- ClonalityPolyclonal
- Concentration0.59 mg/ml
- ConjugateUnconjugated
- Gene ID248
- Target nameALPI
- Target descriptionalkaline phosphatase, intestinal
- Target synonymsIAP, intestinal-type alkaline phosphatase, Kasahara isozyme, alkaline phosphomonoesterase, glycerophosphatase, intestinal alkaline phosphatase
- HostRabbit
- IsotypeIgG
- Protein IDP09923
- Protein NameIntestinal-type alkaline phosphatase
- Scientific DescriptionThere are at least four distinct but related alkaline phosphatases: intestinal, placental, placental-like, and liver/bone/kidney (tissue non-specific). The intestinal alkaline phosphatase gene encodes a digestive brush-border enzyme. This enzyme is upregulated during small intestinal epithelial cell differentiation. [provided by RefSeq]
- ReactivityHuman, Mouse
- Storage Instruction-20°C or -80°C,2°C to 8°C
- UNSPSC41116161






![Non-transfected (–) and transfected (+) 293T whole cell extracts were separated by 10% SDS-PAGE, and the membrane was blotted with Alkaline phosphatase antibody [HL3439] (GTX641312) diluted at 1:5000. The HRP-conjugated anti-rabbit IgG antibody (GTX213110-01) was used to detect the primary antibody.](https://www.genetex.com/upload/website/prouct_img/normal/GTX641312/GTX641312_T-45593_20241129_WB_multiple_B_24120522_789.webp)