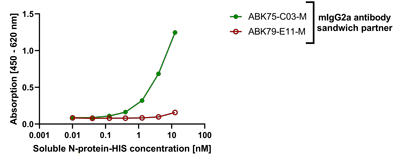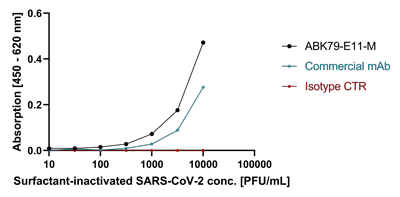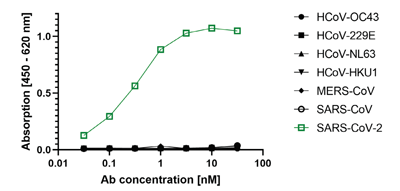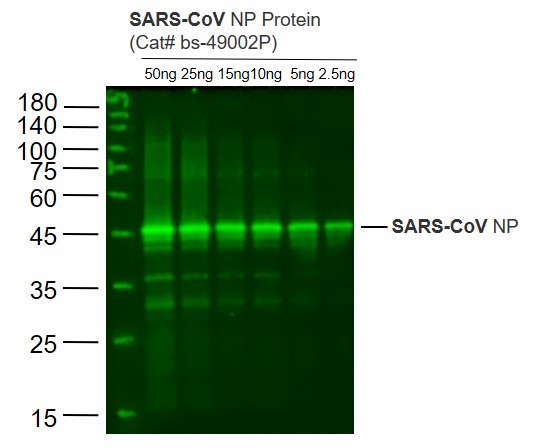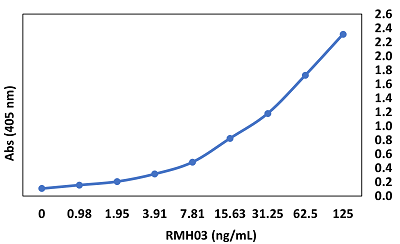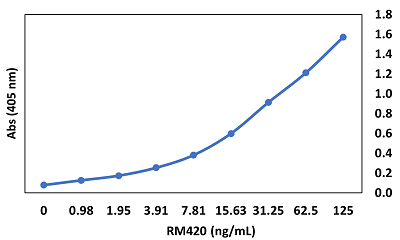
anti-SARS-CoV-2 Nucleocapsid, Rabbit Monoclonal (RM420)
REV-31-1307-00
ApplicationsELISA
Product group Antibodies
ReactivityVirus
TargetN
Overview
- SupplierRevMAb Biosciences
- Product Nameanti-SARS-CoV-2 Nucleocapsid, Rabbit Monoclonal (RM420)
- Delivery Days Customer2
- ApplicationsELISA
- CertificationResearch Use Only
- ClonalityMonoclonal
- Clone IDRM420
- Concentration1 mg/ml
- Gene ID1489678
- Target nameN
- Target descriptionORF9a protein;nucleocapsid protein
- Target synonymssars9a, ORF9a protein;nucleocapsid protein
- HostRabbit
- IsotypeIgG
- Protein IDP59595
- Protein NameNucleoprotein
- Scientific DescriptionRecombinant Antibody. This antibody reacts to 2019-nCov Nucleocapsid protein. Applications: ELISA. Clone: RM420. Isotype: Rabbit IgG. Formulation: Liquid. 50% Glycerol/PBS with 1% BSA and 0.09% sodium azide. SARS-CoV-2 shares 79.5% sequence identity with SARS-CoV and is 96.2% identical at the genome level to the bat coronavirus BatCoV RaTG133, suggesting it had originated in bats. The coronaviral genome encodes four major structural proteins: the Spike (S) protein, Nucleocapsid (N) protein, Membrane/Matrix (M) protein and the Envelope (E) protein. The SARS Envelope (E) protein contains a short palindromic transmembrane helical hairpin that seems to deform lipid bilayers, which may explain its role in viral budding and virion envelope morphogenesis. The SARS Membrane/Matrix (M) protein is one of the major structural viral proteins. It is an integral membrane protein involved in the budding of the viral particles and interacts with SARS Spike (S) protein and the Nucleocapsid (N) protein. The CoV Spike (S) protein assembles as trimer and plays the most important role in viral attachment, fusion and entry. It is composed of a short intracellular tail, a transmembrane anchor and a large ectodomain that consists of a receptor binding S1 subunit (RBD domain) and a membrane-fusing S2 subunit. Sequence analysis of the SARS-CoV-2 S protein genome showed that it was only 75% identical with the SARS-CoV S protein. However, analysis of the receptor binding motif (RBM) in the S protein showed that most of the amino acid residues essential for receptor binding were conserved between SARS-CoV and SARS-CoV-2, suggesting that the 2 CoV strains use the same host receptor for cell entry, the receptor angiotensin-converting enzyme 2 (ACE2). The N protein contains two domains, both of them bind the virus RNA genome via different mechanisms. - SARS-CoV-2 shares 79.5% sequence identity with SARS-CoV and is 96.2% identical at the genome level to the bat coronavirus BatCoV RaTG133, suggesting it had originated in bats. The coronaviral genome encodes four major structural proteins: the Spike (S) protein, Nucleocapsid (N) protein, Membrane/Matrix (M) protein and the Envelope (E) protein. The SARS Envelope (E) protein contains a short palindromic transmembrane helical hairpin that seems to deform lipid bilayers, which may explain its role in viral budding and virion envelope morphogenesis. The SARS Membrane/Matrix (M) protein is one of the major structural viral proteins. It is an integral membrane protein involved in the budding of the viral particles and interacts with SARS Spike (S) protein and the Nucleocapsid (N) protein. The CoV Spike (S) protein assembles as trimer and plays the most important role in viral attachment, fusion and entry. It is composed of a short intracellular tail, a transmembrane anchor and a large ectodomain that consists of a receptor binding S1 subunit (RBD domain) and a membrane-fusing S2 subunit. Sequence analysis of the SARS-CoV-2 S protein genome showed that it was only 75% identical with the SARS-CoV S protein. However, analysis of the receptor binding motif (RBM) in the S protein showed that most of the amino acid residues essential for receptor binding were conserved between SARS-CoV and SARS-CoV-2, suggesting that the 2 CoV strains use the same host receptor for cell entry, the receptor angiotensin-converting enzyme 2 (ACE2). The N protein contains two domains, both of them bind the virus RNA genome via different mechanisms.
- ReactivityVirus
- Storage Instruction-20°C,2°C to 8°C
- UNSPSC41116161