![IHC-P analysis of pancreas tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50 IHC-P analysis of pancreas tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50](https://www.genetex.com/upload/website/prouct_img/normal/GTX84692/GTX84692_3033_IHC-P_w_23061420_360.webp)
IHC-P analysis of pancreas tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50
CUGBP1 antibody [5B8]
GTX84692
ApplicationsImmunoFluorescence, Western Blot, ImmunoCytoChemistry, ImmunoHistoChemistry, ImmunoHistoChemistry Paraffin
Product group Antibodies
ReactivityHuman, Monkey, Mouse
TargetCELF1
Overview
- SupplierGeneTex
- Product NameCUGBP1 antibody [5B8]
- Delivery Days Customer9
- Application Supplier NoteWB: 1:2000. ICC/IF: 1:100. IHC-P: 1:50. *Optimal dilutions/concentrations should be determined by the researcher.Not tested in other applications.
- ApplicationsImmunoFluorescence, Western Blot, ImmunoCytoChemistry, ImmunoHistoChemistry, ImmunoHistoChemistry Paraffin
- CertificationResearch Use Only
- ClonalityMonoclonal
- Clone ID5B8
- Concentration0.99 mg/ml
- ConjugateUnconjugated
- Gene ID10658
- Target nameCELF1
- Target descriptionCUGBP Elav-like family member 1
- Target synonymsBRUNOL2, CUG-BP, CUGBP, CUGBP1, EDEN-BP, NAB50, NAPOR, hNab50, CUGBP Elav-like family member 1, 50 kDa nuclear polyadenylated RNA-binding protein, CUG RNA-binding protein, CUG triplet repeat RNA-binding protein 1, CUG-BP- and ETR-3-like factor 1, EDEN-BP homolog, RNA-binding protein BRUNOL-2, bruno-like 2, bruno-like protein 2, deadenylation factor CUG-BP, embryo deadenylation element binding protein, embryo deadenylation element-binding protein homolog, nuclear polyadenylated RNA-binding protein, 50-kD
- HostMouse
- IsotypeIgG2b
- Protein IDQ92879
- Protein NameCUGBP Elav-like family member 1
- Scientific DescriptionRNA-binding protein implicated in the regulation of several post-transcriptional events. Involved in pre-mRNA alternative splicing, mRNA translation and stability. Mediates exon inclusion and/or exclusion in pre-mRNA that are subject to tissue-specific and developmentally regulated alternative splicing. Specifically activates exon 5 inclusion of cardiac isoforms of TNNT2 during heart remodeling at the juvenile to adult transition. Acts as both an activator and repressor of a pair of coregulated exons: promotes inclusion of the smooth muscle (SM) exon but exclusion of the non-muscle (NM) exon in actinin pre-mRNAs. Activates SM exon 5 inclusion by antagonizing the repressive effect of PTB. Promotes exclusion of exon 11 of the INSR pre-mRNA. Inhibits, together with HNRNPH1, insulin receptor (IR) pre-mRNA exon 11 inclusion in myoblast. Increases translation and controls the choice of translation initiation codon of CEBPB mRNA. Increases mRNA translation of CEBPB in aging liver (By similarity). Increases translation of CDKN1A mRNA by antagonizing the repressive effect of CALR3. Mediates rapid cytoplasmic mRNA deadenylation. Recruits the deadenylase PARN to the poly(A) tail of EDEN-containing mRNAs to promote their deadenylation. Required for completion of spermatogenesis (By similarity). Binds to (CUG)n triplet repeats in the 3-UTR of transcripts such as DMPK and to Bruno response elements (BREs). Binds to muscle-specific splicing enhancer (MSE) intronic sites flanking the alternative exon 5 of TNNT2 pre-mRNA. Binds to AU-rich sequences (AREs or EDEN-like) localized in the 3-UTR of JUN and FOS mRNAs. Binds to the IR RNA. Binds to the 5-region of CDKN1A and CEBPB mRNAs. Binds with the 5-region of CEBPB mRNA in aging liver.
- ReactivityHuman, Monkey, Mouse
- Storage Instruction-20°C or -80°C,2°C to 8°C
- UNSPSC41116161

![IHC-P analysis of prostate carcinoma tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50 IHC-P analysis of prostate carcinoma tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50](https://www.genetex.com/upload/website/prouct_img/normal/GTX84692/GTX84692_3030_IHC-P_w_23061420_950.webp)
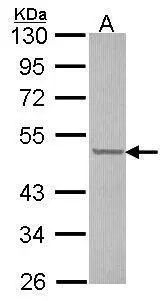
![WB analysis of various cell lines using GTX84692 CUGBP1 antibody [5B8]. Loading : 10 ug per lane Dilution : 1:200 WB analysis of various cell lines using GTX84692 CUGBP1 antibody [5B8]. Loading : 10 ug per lane Dilution : 1:200](https://www.genetex.com/upload/website/prouct_img/normal/GTX84692/GTX84692_3560_WB_w_23061420_375.webp)
![IHC-P analysis of kidney tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50 IHC-P analysis of kidney tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50](https://www.genetex.com/upload/website/prouct_img/normal/GTX84692/GTX84692_3031_IHC-P_w_23061420_414.webp)
![WB analysis of various samples using GTX84692 CUGBP1 antibody [5B8]. Loading : 10 ug per lane Dilution : 1:200 WB analysis of various samples using GTX84692 CUGBP1 antibody [5B8]. Loading : 10 ug per lane Dilution : 1:200](https://www.genetex.com/upload/website/prouct_img/normal/GTX84692/GTX84692_3561_WB_w_23061420_890.webp)
![ICC/IF analysis of COS7 cells transiently transfected with CUGBP1 plasmid using GTX84692 CUGBP1 antibody [5B8]. ICC/IF analysis of COS7 cells transiently transfected with CUGBP1 plasmid using GTX84692 CUGBP1 antibody [5B8].](https://www.genetex.com/upload/website/prouct_img/normal/GTX84692/GTX84692_1192_ICCIF_w_23061420_811.webp)
![IHC-P analysis of colon adenocarcinoma tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50 IHC-P analysis of colon adenocarcinoma tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50](https://www.genetex.com/upload/website/prouct_img/normal/GTX84692/GTX84692_3028_IHC-P_w_23061420_531.webp)
![IHC-P analysis of liver tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50 IHC-P analysis of liver tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50](https://www.genetex.com/upload/website/prouct_img/normal/GTX84692/GTX84692_3032_IHC-P_w_23061420_933.webp)
![IHC-P analysis of human ovary adenocarcinoma tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50 IHC-P analysis of human ovary adenocarcinoma tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50](https://www.genetex.com/upload/website/prouct_img/normal/GTX84692/GTX84692_3029_IHC-P_w_23061420_526.webp)
![IHC-P analysis of prostate tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50 IHC-P analysis of prostate tissue using GTX84692 CUGBP1 antibody [5B8]. Antigen retrieval : Heat-induced epitope retrieval by 10mM citrate buffer, pH6.0, 100oC for 10min. Dilution : 1:50](https://www.genetex.com/upload/website/prouct_img/normal/GTX84692/GTX84692_3034_IHC-P_w_23061420_858.webp)





![ICC/IF analysis of normal and DM1 (dystrophia myotonica) myoblasts using GTX29549 CUGBP1 antibody [3B1].](https://www.genetex.com/upload/website/prouct_img/normal/GTX29549/GTX29549_172_ICC-IF_w_23060722_920.webp)