ELISA Kit for Acid Sphingomyelinase (ASM)
SEB360HU
Product group Assays
Overview
- SupplierCloud-Clone Corp.
- Product NameELISA Kit for Acid Sphingomyelinase (ASM)
- Delivery Days Customer12
- ApplicationsELISA
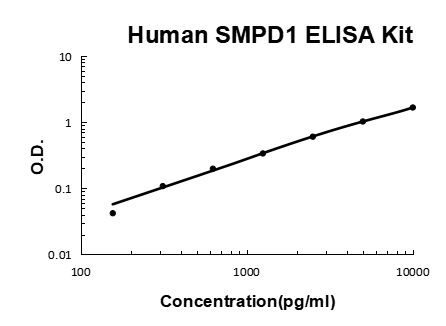
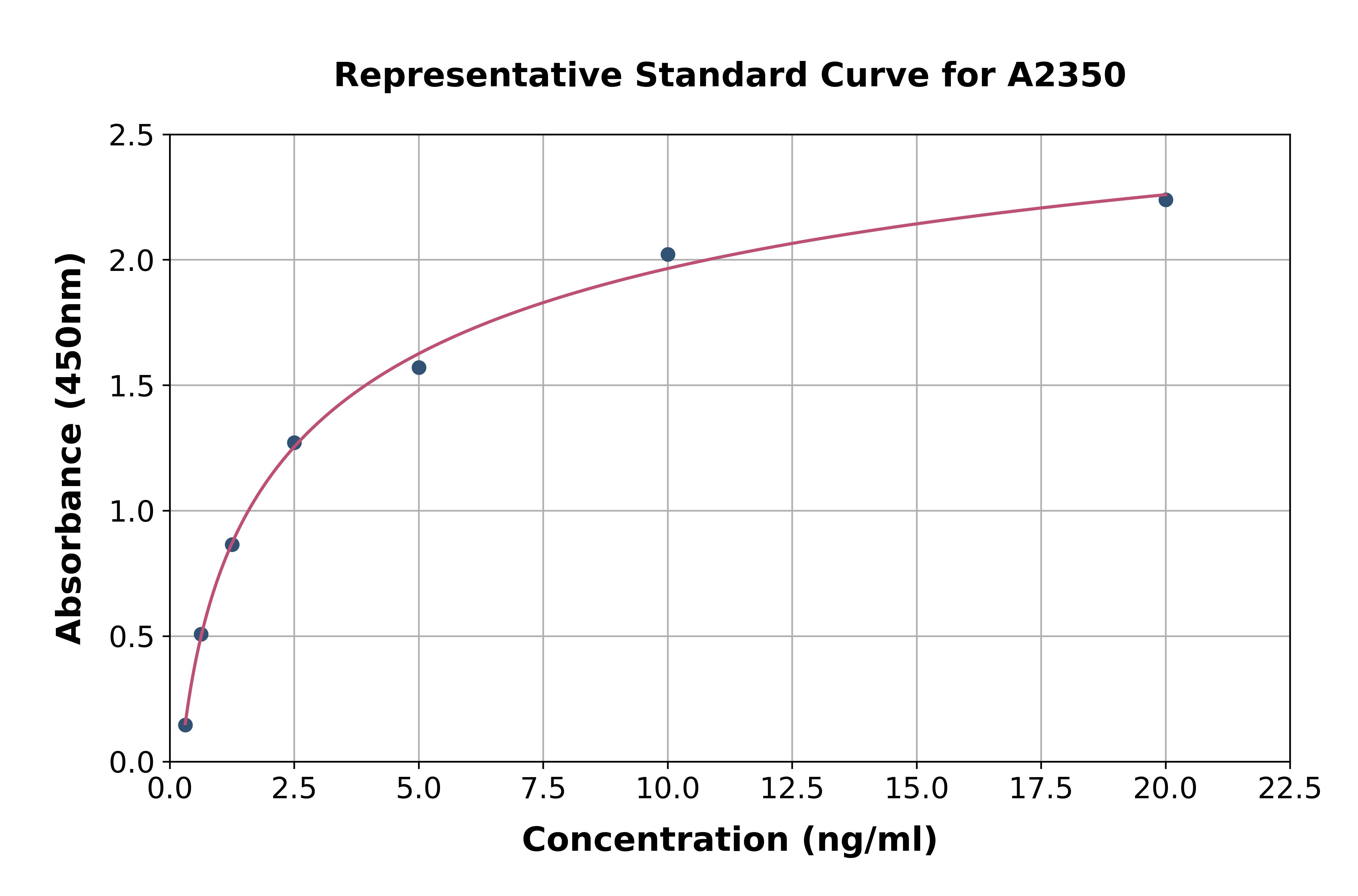
- Assay Detection Range0.156-10ng/mL
- Assay PrecisionIntra-assay Precision (Precision within an assay): 3 samples with low, middle and high level Acid Sphingomyelinase (ASM) were tested 20 times on one plate, respectively. Inter-assay Precision (Precision between assays): 3 samples with low, middle...
- Assay SensitivityThe minimum detectable dose of this kit is typically less than 0.057ng/mL
- Assay Test PrincipleThe test principle applied in this kit is Sandwich enzyme immunoassay. The microtiter plate provided in this kit has been pre-coated with an antibody specific to Acid Sphingomyelinase (ASM). Standards or samples are then added to the appropriate...
- Assay Time3h
- CertificationResearch Use Only
- Protein IDP17405
- Protein NameSphingomyelin phosphodiesterase
- UNSPSC41116100
- SpeciesHuman
References
- Evaluation of sphingolipid metabolism on diabetic retinopathy. Ensari Delioglu EN et al., 2021 Nov, Indian J OphthalmolRead this paper
- Acid Sphingomyelinase and Acid beta-Glucosidase 1 Exert Opposite Effects on Interleukin-1beta-Induced Interleukin 6 Production in Rheumatoid Arthritis Fibroblast-Like Synoviocytes. Zhao M et al., 2021 Aug, InflammationRead this paper
- Fenofibrate decreases plasma ceramide in type 2 diabetes patients: A novel marker of CVD? Croyal M et al., 2018 Mar, Diabetes MetabRead this paper
- Acid sphingomyelinase is activated in sickle cell erythrocytes and contributes to inflammatory microparticle generation in SCD. Awojoodu AO et al., 2014 Sep 18, BloodRead this paper