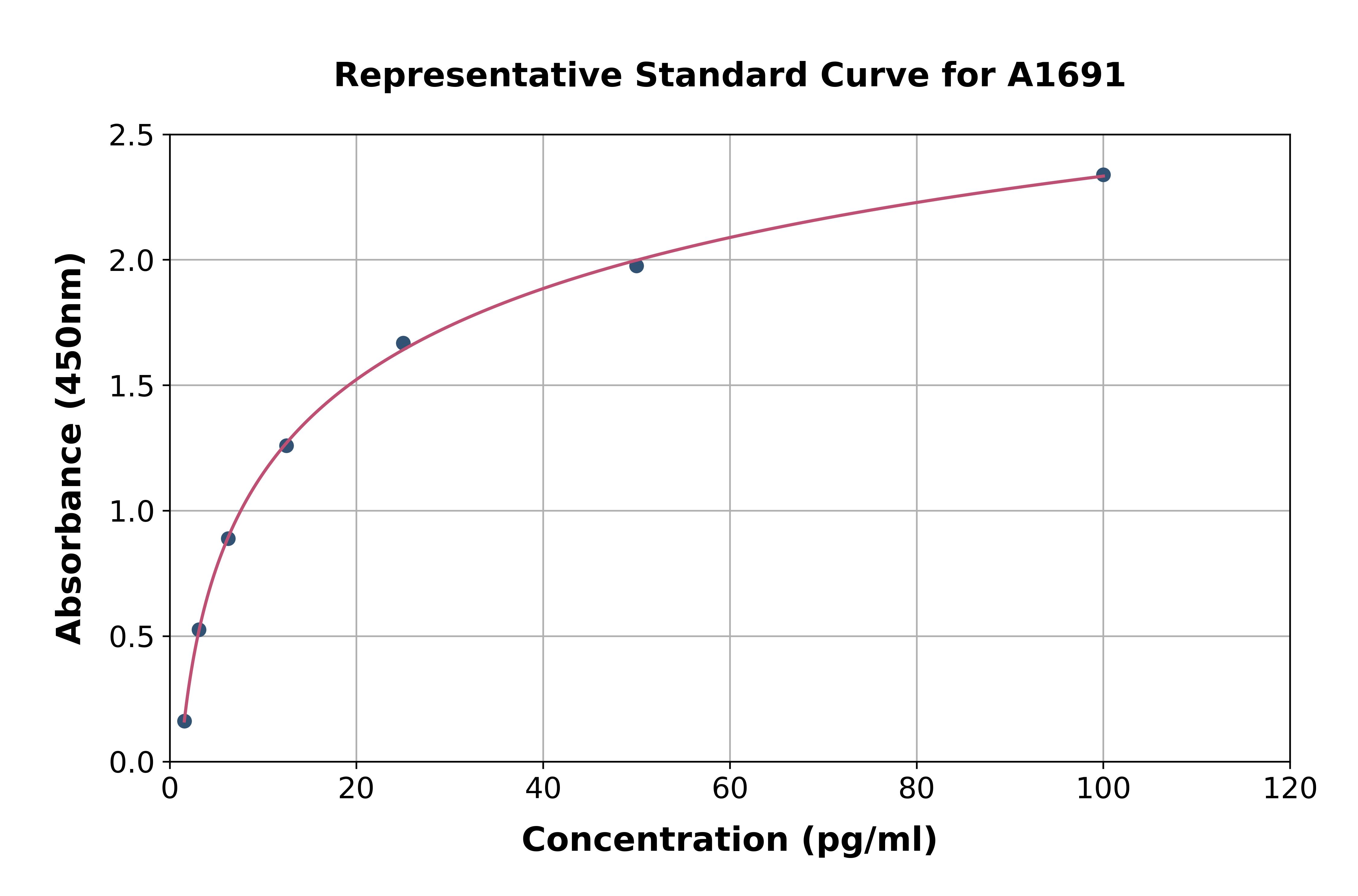
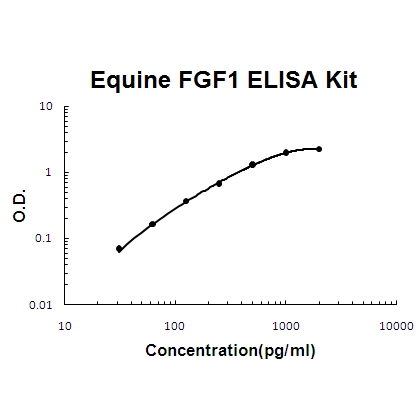
Human FGF1 PicoKine ELISA Kit standard curve
Human FGF1/Fgf Acidic ELISA Kit PicoKine(r)
EK0339
Product group Assays
Overview
- SupplierBoster Bio
- Product NameHuman FGF1 PicoKine ELISA Kit
- Delivery Days Customer9
- ApplicationsELISA
- Assay Detection Range31.2 pg/ml - 2,000 pg/ml
- Assay Sensitivity<10 pg/ml
- CertificationResearch Use Only
- Scientific DescriptionHuman FGF1/Fgf Acidic ELISA Kit PicoKine® (96 Tests). Quantitate Human FGF1 in cell culture supernatants, cell lysates, serum and plasma (heparin, EDTA). Sensitivity: 10pg/ml. The brand Picokine indicates this is a premium quality ELISA kit. Each Picokine kit delivers precise quantification, high sensitivity, and excellent reproducibility. Only our most reliable and effective kits qualify as Picokine, guaranteeing top-tier results for your assays.
- Storage Instruction-20°C,2°C to 8°C
- UNSPSC41116158
References
- Zhang J, Xiang Y, Yang Q, et al. Adipose-derived stem cells derived decellularized extracellular matrix enabled skin regeneration and remodeling. Front Bioeng Biotechnol. 2024,12:1347995. doi: 10.3389/fbioe.2024.1347995Read this paper
- Cheng C, Wang J, Xu P, et al. Gremlin1 is a therapeutically targetable FGFR1 ligand that regulates lineage plasticity and castration resistance in prostate cancer. Nat Cancer. 2022,3(5):565-580. doi: 10.1038/s43018-022-00380-3Read this paper
- Li Z, Fu WJ, Chen XQ, et al. Autophagy-based unconventional secretion of HMGB1 in glioblastoma promotes chemosensitivity to temozolomide through macrophage M1-like polarization. J Exp Clin Cancer Res. 2022,41(1):74. doi: 10.1186/s13046-022-02291-8Read this paper
- Matsuo K. CRISPR/Cas9-mediated knockout of the DCL2 and DCL4 genes in Nicotiana benthamiana and its productivity of recombinant proteins. Plant Cell Rep. 2022,41(2):307-317. doi: 10.1007/s00299-021-02809-yRead this paper
- Chen SY, Wang F, Yan XY, et al. Autologous transplantation of EPCs encoding FGF1 gene promotes neovascularization in a porcine model of chronic myocardial ischemia. Int J Cardiol. 2009,135(2):223-32. doi: 10.1016/j.ijcard.2008.12.193Read this paper