![DNA ligase I antibody [10H5] detects DNA ligase I protein at nucleus by immunohistochemical analysis. Sample: Paraffin-embedded human breast carcinoma. DNA ligase I stained by DNA ligase I antibody [10H5] (GTX70141) diluted at 1:100. Antigen Retrieval: Citrate buffer, pH 6.0, 15 min DNA ligase I antibody [10H5] detects DNA ligase I protein at nucleus by immunohistochemical analysis. Sample: Paraffin-embedded human breast carcinoma. DNA ligase I stained by DNA ligase I antibody [10H5] (GTX70141) diluted at 1:100. Antigen Retrieval: Citrate buffer, pH 6.0, 15 min](https://www.genetex.com/upload/website/prouct_img/normal/GTX70141/GTX70141_44020_20200904_IHC-P_w_23061221_918.webp)
DNA ligase I antibody [10H5] detects DNA ligase I protein at nucleus by immunohistochemical analysis. Sample: Paraffin-embedded human breast carcinoma. DNA ligase I stained by DNA ligase I antibody [10H5] (GTX70141) diluted at 1:100. Antigen Retrieval: Citrate buffer, pH 6.0, 15 min
DNA ligase I antibody [10H5]
GTX70141
ApplicationsImmunoFluorescence, ImmunoPrecipitation, Western Blot, ImmunoCytoChemistry, ImmunoHistoChemistry, ImmunoHistoChemistry Paraffin
Product group Antibodies
ReactivityHuman, Mouse
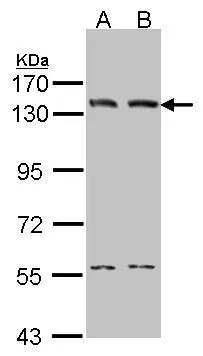
TargetLIG1
Overview
- SupplierGeneTex
- Product NameDNA ligase I antibody [10H5]
- Delivery Days Customer9
- ApplicationsImmunoFluorescence, ImmunoPrecipitation, Western Blot, ImmunoCytoChemistry, ImmunoHistoChemistry, ImmunoHistoChemistry Paraffin
- CertificationResearch Use Only
- ClonalityMonoclonal
- Clone ID10H5
- Concentration0.5 mg/ml
- ConjugateUnconjugated
- Gene ID3978
- Target nameLIG1
- Target descriptionDNA ligase 1
- Target synonymsIMD96, LIGI, hLig1, DNA ligase 1, ligase I, DNA, ATP-dependent, polydeoxyribonucleotide synthase [ATP] 1
- HostMouse
- IsotypeIgG1
- Protein IDP18858
- Protein NameDNA ligase 1
- Scientific DescriptionThis gene encodes a member of the ATP-dependent DNA ligase protein family. The encoded protein functions in DNA replication, recombination, and the base excision repair process. Mutations in this gene that lead to DNA ligase I deficiency result in immunodeficiency and increased sensitivity to DNA-damaging agents. Disruption of this gene may also be associated with a variety of cancers. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2014]
- ReactivityHuman, Mouse
- Storage Instruction-20°C or -80°C,2°C to 8°C
- UNSPSC41116161

![DNA ligase I antibody [10H5] detects DNA ligase I protein at nucleus by immunofluorescent analysis. Sample: HeLa cells were fixed in 4% paraformaldehyde at RT for 15 min. Green: DNA ligase I stained by DNA ligase I antibody [10H5] (GTX70141) diluted at 1:500. Red: phalloidin, a cytoskeleton marker, diluted at 1:200. Scale bar= 10 μm. DNA ligase I antibody [10H5] detects DNA ligase I protein at nucleus by immunofluorescent analysis. Sample: HeLa cells were fixed in 4% paraformaldehyde at RT for 15 min. Green: DNA ligase I stained by DNA ligase I antibody [10H5] (GTX70141) diluted at 1:500. Red: phalloidin, a cytoskeleton marker, diluted at 1:200. Scale bar= 10 μm.](https://www.genetex.com/upload/website/prouct_img/normal/GTX70141/GTX70141_43964_20200722_ICC_IF_w_23061221_777.webp)
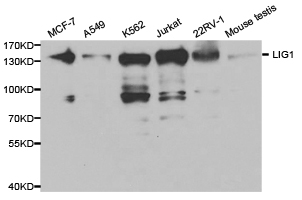
![Various whole cell extracts (30 μg) were separated by 5% SDS-PAGE, and the membrane was blotted with DNA ligase I antibody [10H5] (GTX70141) diluted at 1:2000. The HRP-conjugated anti-mouse IgG antibody (GTX213111-01) was used to detect the primary antibody. Various whole cell extracts (30 μg) were separated by 5% SDS-PAGE, and the membrane was blotted with DNA ligase I antibody [10H5] (GTX70141) diluted at 1:2000. The HRP-conjugated anti-mouse IgG antibody (GTX213111-01) was used to detect the primary antibody.](https://www.genetex.com/upload/website/prouct_img/normal/GTX70141/GTX70141_44020_20200724_WB_23073119_984.webp)