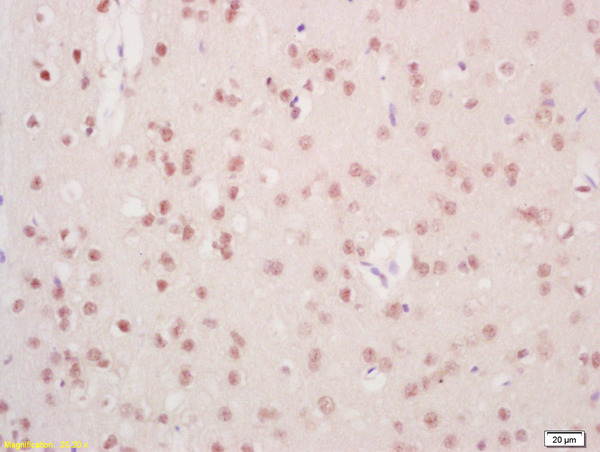
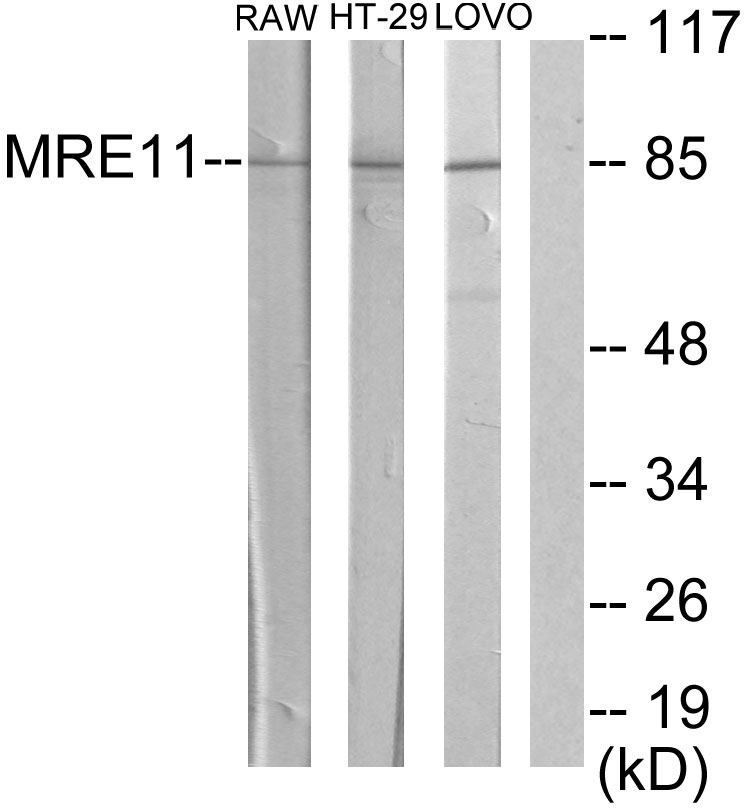
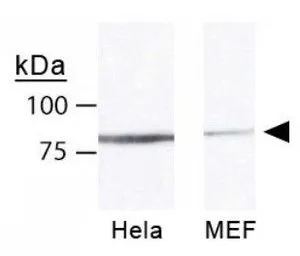
Mre11 antibody [12D7]
GTX70212
ApplicationsFunctional Assay, ImmunoFluorescence, ImmunoPrecipitation, Western Blot, ELISA, ImmunoCytoChemistry, Other Application
Product group Antibodies
ReactivityHuman
TargetMRE11
Overview
- SupplierGeneTex
- Product NameMre11 antibody [12D7]
- Delivery Days Customer9
- Application Supplier NoteWB: 1:500-1:3000. ICC/IF: 1:100-1:1000. *Optimal dilutions/concentrations should be determined by the researcher.Not tested in other applications.
- ApplicationsFunctional Assay, ImmunoFluorescence, ImmunoPrecipitation, Western Blot, ELISA, ImmunoCytoChemistry, Other Application
- CertificationResearch Use Only
- ClonalityMonoclonal
- Clone ID12D7
- Concentration0.45 mg/ml
- ConjugateUnconjugated
- Gene ID4361
- Target nameMRE11
- Target descriptionMRE11 double strand break repair nuclease
- Target synonymsATLD, HNGS1, MRE11A, MRE11B, double-strand break repair protein MRE11, AT-like disease, DNA recombination and repair protein, MRE11 double strand break repair nuclease A, MRE11 homolog 1, MRE11 homolog A, double strand break repair nuclease, MRE11 homolog, double strand break repair nuclease A, MRE11 meiotic recombination 11 homolog A, MRE11 meiotic recombination 11-like protein A, double-strand break repair protein MRE11A, endo/exonuclease Mre11, meiotic recombination 11 homolog 1, meiotic recombination 11 homolog A
- HostMouse
- IsotypeIgG1
- Protein IDP49959
- Protein NameDouble-strand break repair protein MRE11
- Scientific DescriptionThis gene encodes a nuclear protein involved in homologous recombination, telomere length maintenance, and DNA double-strand break repair. By itself, the protein has 3 to 5 exonuclease activity and endonuclease activity. The protein forms a complex with the RAD50 homolog; this complex is required for nonhomologous joining of DNA ends and possesses increased single-stranded DNA endonuclease and 3 to 5 exonuclease activities. In conjunction with a DNA ligase, this protein promotes the joining of noncomplementary ends in vitro using short homologies near the ends of the DNA fragments. This gene has a pseudogene on chromosome 3. Alternative splicing of this gene results in two transcript variants encoding different isoforms. [provided by RefSeq, Jul 2008]
- ReactivityHuman
- Storage Instruction-20°C or -80°C,2°C to 8°C
- UNSPSC12352203
References
- Hale A, Dhoonmoon A, Straka J, et al. Multi-step processing of replication stress-derived nascent strand DNA gaps by MRE11 and EXO1 nucleases. Nat Commun. 2023,14(1):6265. doi: 10.1038/s41467-023-42011-0Read this paper
- Packard JE, Williams MR, Fromuth DP, et al. Proliferating cell nuclear antigen inhibitors block distinct stages of herpes simplex virus infection. PLoS Pathog. 2023,19(7):e1011539. doi: 10.1371/journal.ppat.1011539Read this paper
- Trivedi P, Steele CD, Au FKC, et al. Mitotic tethering enables inheritance of shattered micronuclear chromosomes. Nature. 2023,618(7967):1049-1056. doi: 10.1038/s41586-023-06216-zRead this paper
- Nguyen DD, Kim E, Le NT, et al. Deficiency in mammalian STN1 promotes colon cancer development via inhibiting DNA repair. Sci Adv. 2023,9(19):eadd8023. doi: 10.1126/sciadv.add8023Read this paper
- Saha LK, Saha S, Yang X, et al. Replication-associated formation and repair of human topoisomerase IIIα cleavage complexes. Nat Commun. 2023,14(1):1925. doi: 10.1038/s41467-023-37498-6Read this paper
- Thakar T, Dhoonmoon A, Straka J, et al. Lagging strand gap suppression connects BRCA-mediated fork protection to nucleosome assembly through PCNA-dependent CAF-1 recycling. Nat Commun. 2022,13(1):5323. doi: 10.1038/s41467-022-33028-yRead this paper
- Dhoonmoon A, Nicolae CM, Moldovan GL. The KU-PARP14 axis differentially regulates DNA resection at stalled replication forks by MRE11 and EXO1. Nat Commun. 2022,13(1):5063. doi: 10.1038/s41467-022-32756-5Read this paper
- Ko JMY, Lam SY, Ning L, et al. RAD50 Loss of Function Variants in the Zinc Hook Domain Associated with Higher Risk of Familial Esophageal Squamous Cell Carcinoma. Cancers (Basel). 2021,13(18). doi: 10.3390/cancers13184715Read this paper
- Kurashima K, Kashiwagi H, Shimomura I, et al. SMARCA4 deficiency-associated heterochromatin induces intrinsic DNA replication stress and susceptibility to ATR inhibition in lung adenocarcinoma. NAR Cancer. 2020,2(2):zcaa005. doi: 10.1093/narcan/zcaa005Read this paper
- Xu X, Ni K, He Y, et al. The epigenetic regulator LSH maintains fork protection and genomic stability via MacroH2A deposition and RAD51 filament formation. Nat Commun. 2021,12(1):3520. doi: 10.1038/s41467-021-23809-2Read this paper