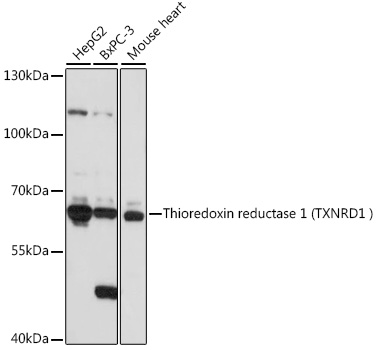
TrxR1 antibody [N3C2], Internal
GTX108727
ApplicationsImmunoFluorescence, Western Blot, ImmunoCytoChemistry, ImmunoHistoChemistry, ImmunoHistoChemistry Paraffin
Product group Antibodies
ReactivityHuman, Mouse, Rat
TargetTXNRD1
Overview
- SupplierGeneTex
- Product NameTrxR1 antibody [N3C2], Internal
- Delivery Days Customer9
- Application Supplier NoteWB: 1:500-1:3000. ICC/IF: 1:100-1:1000. IHC-P: 1:100-1:1000. *Optimal dilutions/concentrations should be determined by the researcher.Not tested in other applications.
- ApplicationsImmunoFluorescence, Western Blot, ImmunoCytoChemistry, ImmunoHistoChemistry, ImmunoHistoChemistry Paraffin
- CertificationResearch Use Only
- ClonalityPolyclonal
- Concentration1 mg/ml
- ConjugateUnconjugated
- Gene ID7296
- Target nameTXNRD1
- Target descriptionthioredoxin reductase 1
- Target synonymsGRIM-12, TR, TR1, TRXR1, TXNR, TXNR1, thioredoxin reductase 1, cytoplasmic, KM-102-derived reductase-like factor, gene associated with retinoic and IFN-induced mortality 12 protein, gene associated with retinoic and interferon-induced mortality 12 protein, oxidoreductase, peroxidase TXNRD1, selenoprotein TXNRD1, testis tissue sperm-binding protein Li 46a, thioredoxin reductase GRIM-12, thioredoxin reductase TR1
- HostRabbit
- IsotypeIgG
- Protein IDQ16881
- Protein NameThioredoxin reductase 1, cytoplasmic
- Scientific DescriptionThis gene encodes a member of the family of pyridine nucleotide oxidoreductases. This protein reduces thioredoxins as well as other substrates, and plays a role in selenium metabolism and protection against oxidative stress. The functional enzyme is thought to be a homodimer which uses FAD as a cofactor. Each subunit contains a selenocysteine (Sec) residue which is required for catalytic activity. The selenocysteine is encoded by the UGA codon that normally signals translation termination. The 3 UTR of selenocysteine-containing genes have a common stem-loop structure, the sec insertion sequence (SECIS), that is necessary for the recognition of UGA as a Sec codon rather than as a stop signal. Alternative splicing results in several transcript variants encoding the same or different isoforms. [provided by RefSeq]
- ReactivityHuman, Mouse, Rat
- Storage Instruction-20°C or -80°C,2°C to 8°C
- UNSPSC12352203
References
- Brinks R, Wruck CJ, Schmitz J, et al. Nrf2 Activation Does Not Protect from Aldosterone-Induced Kidney Damage in Mice. Antioxidants (Basel). 2023,12(3). doi: 10.3390/antiox12030777Read this paper
- Yoon JH, Youn K, Jun M. Protective effect of sargahydroquinoic acid against Aβ(25-35)-evoked damage via PI3K/Akt mediated Nrf2 antioxidant defense system. Biomed Pharmacother. 2021,144:112271. doi: 10.1016/j.biopha.2021.112271Read this paper
- Balhorn R, Hartmann C, Schupp N. Aldosterone Induces DNA Damage and Activation of Nrf2 Mainly in Tubuli of Mouse Kidneys. Int J Mol Sci. 2020,21(13). doi: 10.3390/ijms21134679Read this paper
- Gao Q, Zhang G, Zheng Y, et al. SLC27A5 deficiency activates NRF2/TXNRD1 pathway by increased lipid peroxidation in HCC. Cell Death Differ. 2020,27(3):1086-1104. doi: 10.1038/s41418-019-0399-1Read this paper
- Zimnol A, Amann K, Mandel P, et al. Angiotensin II type 1a receptor-deficient mice develop angiotensin II-induced oxidative stress and DNA damage without blood pressure increase. Am J Physiol Renal Physiol. 2017,313(6):F1264-F1273. doi: 10.1152/ajprenal.00183.2017Read this paper
- Cinghu S, Yellaboina S, Freudenberg JM, et al. Integrative framework for identification of key cell identity genes uncovers determinants of ES cell identity and homeostasis. Proc Natl Acad Sci U S A. 2014,111(16):E1581-90. doi: 10.1073/pnas.1318598111Read this paper
- Queisser N, Oteiza PI, Link S, et al. Aldosterone activates transcription factor Nrf2 in kidney cells both in vitro and in vivo. Antioxid Redox Signal. 2014,21(15):2126-42. doi: 10.1089/ars.2013.5565Read this paper
- Tena-Suck ML, Hernández-Campos ME, Ortiz-Plata A, et al. Intracerebral injection of oil cyst content of human craniopharyngioma (oil machinery fluid) as a toxic model in the rat brain. Acta Histochem. 2014,116(3):448-56. doi: 10.1016/j.acthis.2013.10.002Read this paper