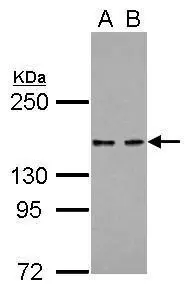
![Various whole cell extracts (30 μg) were separated by 5% SDS-PAGE, and the membrane was blotted with KDM6A antibody [N2C1], Internal (GTX121246) diluted at 1:1000. The HRP-conjugated anti-rabbit IgG antibody (GTX213110-01) was used to detect the primary antibody. Corresponding RNA expression data for the same cell lines are based on Human Protein Atlas program. Various whole cell extracts (30 μg) were separated by 5% SDS-PAGE, and the membrane was blotted with KDM6A antibody [N2C1], Internal (GTX121246) diluted at 1:1000. The HRP-conjugated anti-rabbit IgG antibody (GTX213110-01) was used to detect the primary antibody. Corresponding RNA expression data for the same cell lines are based on Human Protein Atlas program.](https://www.genetex.com/upload/website/prouct_img/normal/GTX121246/GTX121246_42992_20221209_WB_TPM_watermark_22121123_237.webp)
Various whole cell extracts (30 μg) were separated by 5% SDS-PAGE, and the membrane was blotted with KDM6A antibody [N2C1], Internal (GTX121246) diluted at 1:1000. The HRP-conjugated anti-rabbit IgG antibody (GTX213110-01) was used to detect the primary antibody. Corresponding RNA expression data for the same cell lines are based on Human Protein Atlas program.
KDM6A antibody [N2C1], Internal
GTX121246
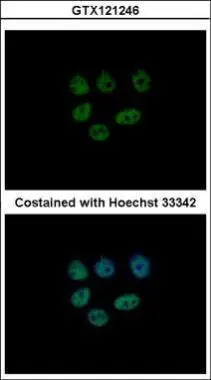
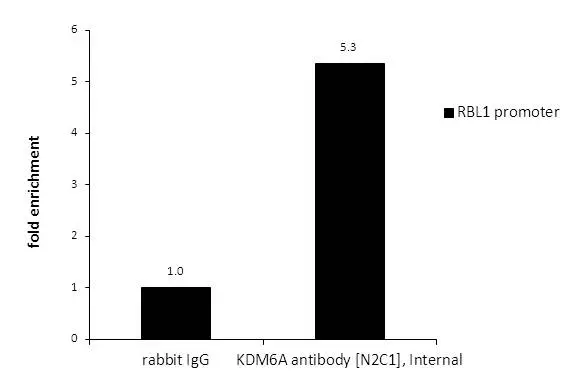
ApplicationsImmunoFluorescence, ImmunoPrecipitation, Western Blot, ChIP Chromatin ImmunoPrecipitation, ImmunoCytoChemistry, ImmunoHistoChemistry, ImmunoHistoChemistry Paraffin
Product group Antibodies
ReactivityHuman, Mouse
TargetKDM6A
Overview
- SupplierGeneTex
- Product NameKDM6A antibody [N2C1], Internal
- Delivery Days Customer9
- Application Supplier NoteWB: 1:500-1:3000. ICC/IF: 1:100-1:1000. IHC-P: 1:100-1:1000. IP: 1:100-1:500. *Optimal dilutions/concentrations should be determined by the researcher.Not tested in other applications.
- ApplicationsImmunoFluorescence, ImmunoPrecipitation, Western Blot, ChIP Chromatin ImmunoPrecipitation, ImmunoCytoChemistry, ImmunoHistoChemistry, ImmunoHistoChemistry Paraffin
- CertificationResearch Use Only
- ClonalityPolyclonal
- Concentration0.78 mg/ml
- ConjugateUnconjugated
- Gene ID7403
- Target nameKDM6A
- Target descriptionlysine demethylase 6A
- Target synonymsKABUK2, UTX, bA386N14.2, lysine-specific demethylase 6A, [histone H3]-trimethyl-L-lysine(27) demethylase 6A, bA386N14.2 (ubiquitously transcribed X chromosome tetratricopeptide repeat protein (UTX)), histone demethylase UTX, lysine (K)-specific demethylase 6A, ubiquitously transcribed tetratricopeptide repeat protein X-linked, ubiquitously-transcribed TPR gene on the X chromosome
- HostRabbit
- IsotypeIgG
- Protein IDO15550
- Protein NameLysine-specific demethylase 6A
- Scientific DescriptionThis gene is located on the X chromosome and is the corresponding locus to a Y-linked gene which encodes a tetratricopeptide repeat (TPR) protein. The encoded protein of this gene contains a JmjC-domain and catalyzes the demethylation of tri/dimethylated histone H3. [provided by RefSeq]
- ReactivityHuman, Mouse
- Storage Instruction-20°C or -80°C,2°C to 8°C
- UNSPSC41116161

![KDM6A antibody [N2C1], Internal immunoprecipitates KDM6A protein in IP experiments. IP samples: SK-N-SH whole cell extract A. 30 μg SK-N-SH whole cell extract B. Control with 4 μg of preimmune Rabbit IgG C. Immunoprecipitation of KDM6A protein by 4 μg KDM6A antibody [N2C1], Internal (GTX121246) 5 % SDS-PAGE The immunoprecipitated KDM6A protein was detected by KDM6A antibody [N2C1], Internal (GTX121246) diluted at 1:500. [EasyBlot anti-rabbit IgG (GTX221666-01) was used as a secondary reagent] KDM6A antibody [N2C1], Internal immunoprecipitates KDM6A protein in IP experiments. IP samples: SK-N-SH whole cell extract A. 30 μg SK-N-SH whole cell extract B. Control with 4 μg of preimmune Rabbit IgG C. Immunoprecipitation of KDM6A protein by 4 μg KDM6A antibody [N2C1], Internal (GTX121246) 5 % SDS-PAGE The immunoprecipitated KDM6A protein was detected by KDM6A antibody [N2C1], Internal (GTX121246) diluted at 1:500. [EasyBlot anti-rabbit IgG (GTX221666-01) was used as a secondary reagent]](https://www.genetex.com/upload/website/prouct_img/normal/GTX121246/GTX121246_40464_IP_w_23060519_669.webp)
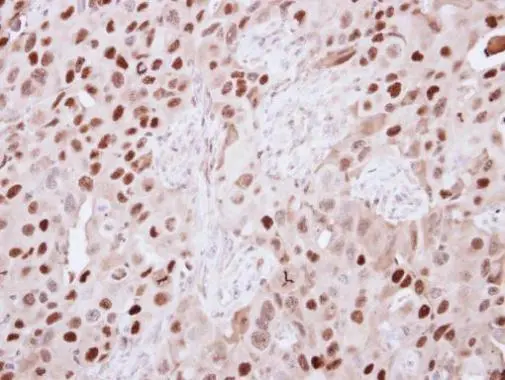
![KDM6A antibody [N2C1], Internal detects KDM6A protein at nucleus by immunohistochemical analysis. Sample: Paraffin-embedded mouse spleen. KDM6A stained by KDM6A antibody [N2C1], Internal (GTX121246) diluted at 1:500. Antigen Retrieval: Citrate buffer, pH 6.0, 15 min KDM6A antibody [N2C1], Internal detects KDM6A protein at nucleus by immunohistochemical analysis. Sample: Paraffin-embedded mouse spleen. KDM6A stained by KDM6A antibody [N2C1], Internal (GTX121246) diluted at 1:500. Antigen Retrieval: Citrate buffer, pH 6.0, 15 min](https://www.genetex.com/upload/website/prouct_img/normal/GTX121246/GTX121246_43054_20180928_IHC-P_M_w_23060519_859.webp)
![Non-transfected (–) and transfected (+) U87-MG whole cell extracts (60 μg) were separated by 5% SDS-PAGE, and the membrane was blotted with KDM6A antibody [N2C1], Internal (GTX121246) diluted at 1:1000. The HRP-conjugated anti-rabbit IgG antibody (GTX213110-01) was used to detect the primary antibody. Non-transfected (–) and transfected (+) U87-MG whole cell extracts (60 μg) were separated by 5% SDS-PAGE, and the membrane was blotted with KDM6A antibody [N2C1], Internal (GTX121246) diluted at 1:1000. The HRP-conjugated anti-rabbit IgG antibody (GTX213110-01) was used to detect the primary antibody.](https://www.genetex.com/upload/website/prouct_img/normal/GTX121246/GTX121246_43055_20181005_WB_shRNA_watermark_w_23060519_683.webp)
![KDM6A antibody [N2C1], Internal detects KDM6A protein at nucleus by immunohistochemical analysis. Sample: Paraffin-embedded mouse lymph node. KDM6A stained by KDM6A antibody [N2C1], Internal (GTX121246) diluted at 1:500. Antigen Retrieval: Citrate buffer, pH 6.0, 15 min KDM6A antibody [N2C1], Internal detects KDM6A protein at nucleus by immunohistochemical analysis. Sample: Paraffin-embedded mouse lymph node. KDM6A stained by KDM6A antibody [N2C1], Internal (GTX121246) diluted at 1:500. Antigen Retrieval: Citrate buffer, pH 6.0, 15 min](https://www.genetex.com/upload/website/prouct_img/normal/GTX121246/GTX121246_43055_20180928_IHC-P_M_w_23060519_785.webp)
![KDM6A antibody [N2C1], Internal detects KDM6A protein at nucleus on mouse duodenum by immunohistochemical analysis. Sample: Paraffin-embedded mouse duodenum. KDM6A antibody [N2C1], Internal (GTX121246) dilution: 1:1000.
Antigen Retrieval: Trilogy? (EDTA based, pH 8.0) buffer, 15min KDM6A antibody [N2C1], Internal detects KDM6A protein at nucleus on mouse duodenum by immunohistochemical analysis. Sample: Paraffin-embedded mouse duodenum. KDM6A antibody [N2C1], Internal (GTX121246) dilution: 1:1000.
Antigen Retrieval: Trilogy? (EDTA based, pH 8.0) buffer, 15min](https://www.genetex.com/upload/website/prouct_img/normal/GTX121246/GTX121246_41563_IHC_M_w_23060519_106.webp)
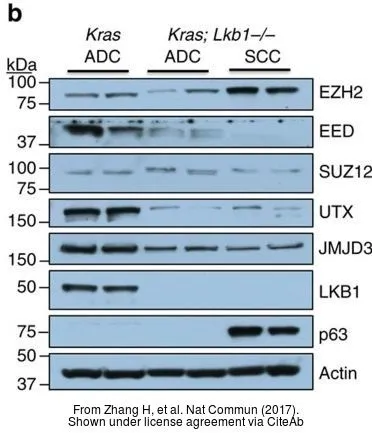



![Various whole cell extracts (30 μg) were separated by 5% SDS-PAGE, and the membrane was blotted with KDM6A antibody [HL1731] (GTX637379) diluted at 1:1000. The HRP-conjugated anti-rabbit IgG antibody (GTX213110-01) was used to detect the primary antibody.](https://www.genetex.com/upload/website/prouct_img/normal/GTX637379/GTX637379_44830_20221028_WB_22110201_396.webp)
![Various whole cell extracts (50 μg) was separated by 5% SDS-PAGE, and the membrane was blotted with KDM6A antibody [HL2068] (GTX637972) diluted at 1:1000. The HRP-conjugated anti-rabbit IgG antibody (GTX213110-01) was used to detect the primary antibody.](https://www.genetex.com/upload/website/prouct_img/normal/GTX637972/GTX637972_T-44886_20221223_WB_M_22122722_423.webp)
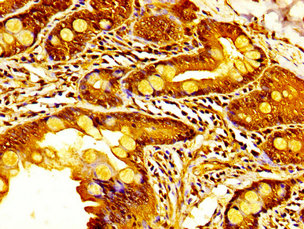
![IHC-P analysis of human squamous cell carcinoma (SCC) from penile tissue using GTX639935 KDM6A antibody [HMV311] HistoMAX?. Squamous cell carcinoma with strong KDM6A staining of tumor cells.](https://www.genetex.com/upload/website/prouct_img/normal/GTX639935/GTX639935_20240403_IHC-P_2_24040301_816.webp)