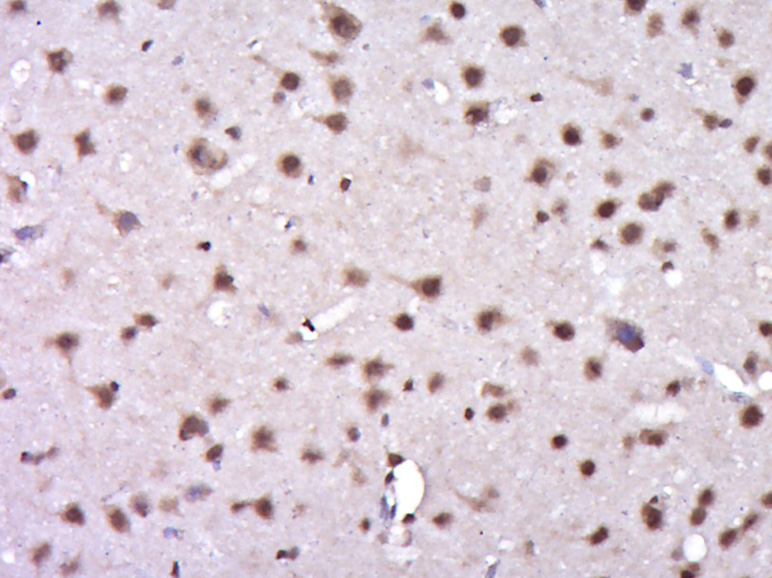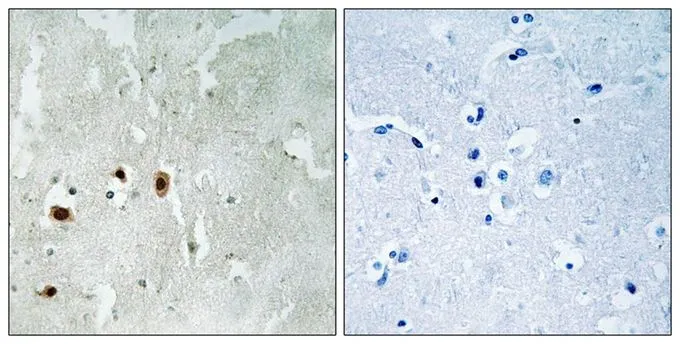
IHC-P analysis of human brain tissue using GTX55370 TIP60 (phospho Ser90) antibody. Left : Primary antibody Right : Primary antibody pre-incubated with the antigen specific peptide
TIP60 (phospho Ser90) antibody
GTX55370
ApplicationsWestern Blot, ImmunoHistoChemistry, ImmunoHistoChemistry Paraffin
Product group Antibodies
ReactivityHuman
TargetKAT5
Overview
- SupplierGeneTex
- Product NameTIP60 (phospho Ser90) antibody
- Delivery Days Customer9
- Application Supplier NoteIHC-P: 1:50-1:100. *Optimal dilutions/concentrations should be determined by the researcher.Not tested in other applications.
- ApplicationsWestern Blot, ImmunoHistoChemistry, ImmunoHistoChemistry Paraffin
- CertificationResearch Use Only
- ClonalityPolyclonal
- Concentration1 mg/ml
- ConjugateUnconjugated
- Gene ID10524
- Target nameKAT5
- Target descriptionlysine acetyltransferase 5
- Target synonymsESA1, HTATIP, HTATIP1, NEDFASB, PLIP, TIP, TIP60, ZC2HC5, cPLA2, histone acetyltransferase KAT5, 60 kDa Tat-interactive protein, HIV-1 Tat interactive protein, 60kDa, K(lysine) acetyltransferase 5, K-acetyltransferase 5, Tat interacting protein, 60kDa, cPLA(2)-interacting protein, cPLA2 interacting protein, histone acetyltransferase HTATIP, protein 2-hydroxyisobutyryltransferase KAT5, protein acetyltransferase KAT5, protein crotonyltransferase KAT5, protein lactyltransferase KAT5
- HostRabbit
- IsotypeIgG
- Protein IDQ92993
- Protein NameHistone acetyltransferase KAT5
- Scientific DescriptionThe protein encoded by this gene belongs to the MYST family of histone acetyl transferases (HATs) and was originally isolated as an HIV-1 TAT-interactive protein. HATs play important roles in regulating chromatin remodeling, transcription and other nuclear processes by acetylating histone and nonhistone proteins. This protein is a histone acetylase that has a role in DNA repair and apoptosis and is thought to play an important role in signal transduction. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Jul 2008]
- ReactivityHuman
- Storage Instruction-20°C or -80°C,2°C to 8°C
- UNSPSC12352203
References
- Li ML, Jiang Q, Bhanu NV, et al. Phosphorylation of TIP60 Suppresses 53BP1 Localization at DNA Damage Sites. Mol Cell Biol. 2019,39(1). doi: 10.1128/MCB.00209-18Read this paper