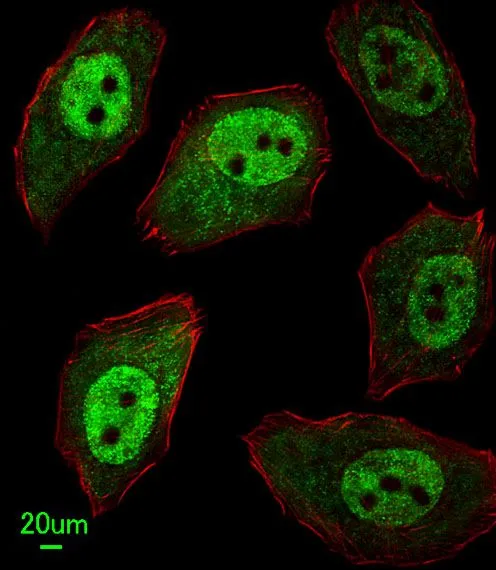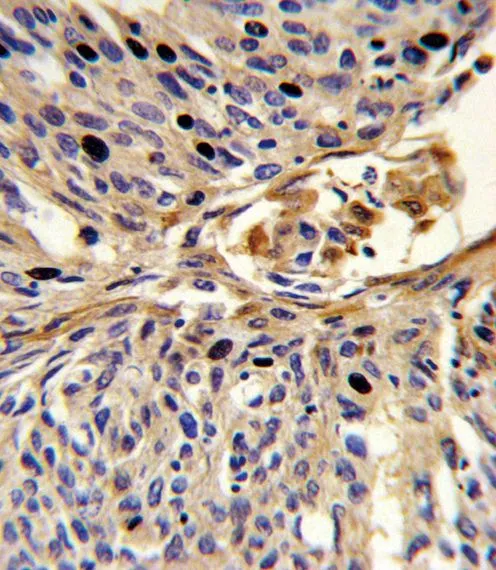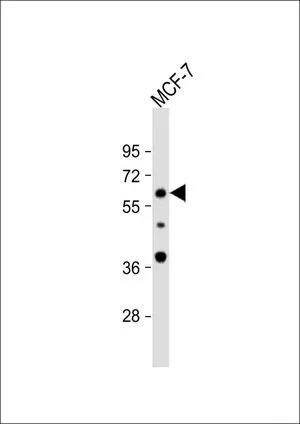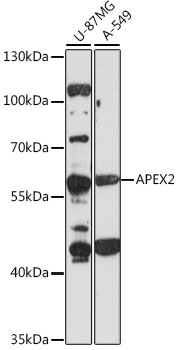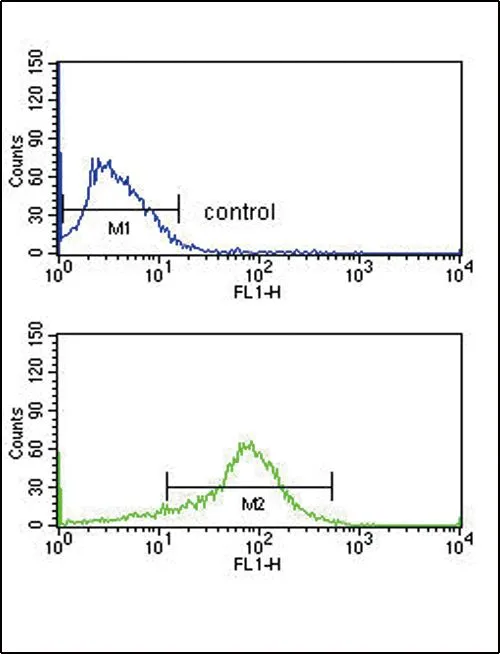
FACS analysis of MCF-7 cells using GTX80642 APEX2 antibody, Internal. Top histogram : negative control Bottom histogram : MCF-7 cells
APEX2 antibody, Internal
GTX80642
ApplicationsFlow Cytometry, ImmunoFluorescence, Western Blot, ImmunoCytoChemistry, ImmunoHistoChemistry, ImmunoHistoChemistry Paraffin
Product group Antibodies
ReactivityHuman
TargetAPEX2
Overview
- SupplierGeneTex
- Product NameAPEX2 antibody, Internal
- Delivery Days Customer9
- Application Supplier NoteWB: 1:1000. ICC/IF: 1:25. IHC-P: 1:10-1:50. FCM: 1:10-1:50. *Optimal dilutions/concentrations should be determined by the researcher.Not tested in other applications.
- ApplicationsFlow Cytometry, ImmunoFluorescence, Western Blot, ImmunoCytoChemistry, ImmunoHistoChemistry, ImmunoHistoChemistry Paraffin
- CertificationResearch Use Only
- ClonalityPolyclonal
- ConjugateUnconjugated
- Gene ID27301
- Target nameAPEX2
- Target descriptionapurinic/apyrimidinic endodeoxyribonuclease 2
- Target synonymsAPE2, APEXL2, XTH2, ZGRF2, DNA-(apurinic or apyrimidinic site) endonuclease 2, AP endonuclease 2, AP endonuclease XTH2, APEX nuclease (apurinic/apyrimidinic endonuclease) 2, DNA-(apurinic or apyrimidinic site) lyase 2, apurinic/apyrimidinic endonuclease-like 2, zinc finger, GRF-type containing 2
- HostRabbit
- IsotypeIgG
- Protein IDQ9UBZ4
- Protein NameDNA-(apurinic or apyrimidinic site) endonuclease 2
- Scientific DescriptionApurinic/apyrimidinic (AP) sites occur frequently in DNA molecules by spontaneous hydrolysis, by DNA damaging agents or by DNA glycosylases that remove specific abnormal bases. AP sites are pre-mutagenic lesions that can prevent normal DNA replication so the cell contains systems to identify and repair such sites. Class II AP endonucleases cleave the phosphodiester backbone 5 to the AP site. This gene encodes a protein shown to have a weak class II AP endonuclease activity. Most of the encoded protein is located in the nucleus but some is also present in mitochondria. This protein may play an important role in both nuclear and mitochondrial base excision repair. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Nov 2012]
- ReactivityHuman
- Storage Instruction-20°C or -80°C,2°C to 8°C
- UNSPSC41116161
References
- APE2 Is a General Regulator of the ATR-Chk1 DNA Damage Response Pathway to Maintain Genome Integrity in Pancreatic Cancer Cells. Hossain MA et al., 2021, Front Cell Dev BiolRead this paper