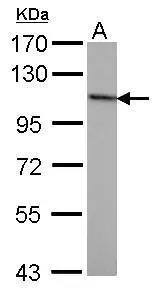
Sample (50 ug of whole cell lysate) A: mouse brain 7.5% SDS PAGE GTX70222 diluted at 1:1000
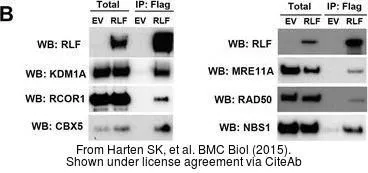
NBS1 antibody [1C3]
GTX70222
ApplicationsImmunoFluorescence, ImmunoPrecipitation, Western Blot, ImmunoCytoChemistry
Product group Antibodies
ReactivityHuman, Mouse
TargetNBN
Overview
- SupplierGeneTex
- Product NameNBS1 antibody [1C3]
- Delivery Days Customer9
- Application Supplier NoteWB: 1:500-1:3000. ICC/IF: 1:100. *Optimal dilutions/concentrations should be determined by the researcher.Not tested in other applications.
- ApplicationsImmunoFluorescence, ImmunoPrecipitation, Western Blot, ImmunoCytoChemistry
- CertificationResearch Use Only
- ClonalityMonoclonal
- Clone ID1C3
- Concentration1 mg/ml
- ConjugateUnconjugated
- Gene ID4683
- Target nameNBN
- Target descriptionnibrin
- Target synonymsAT-V1, AT-V2, ATV, NBS, NBS1, P95, hNbs1, nibrin, Nijmegen breakage syndrome 1 (nibrin), cell cycle regulatory protein p95, p95 protein of the MRE11/RAD50 complex
- HostMouse
- IsotypeIgG1
- Protein IDO60934
- Protein NameNibrin
- Scientific DescriptionMutations in this gene are associated with Nijmegen breakage syndrome, an autosomal recessive chromosomal instability syndrome characterized by microcephaly, growth retardation, immunodeficiency, and cancer predisposition. The encoded protein is a member of the MRE11/RAD50 double-strand break repair complex which consists of 5 proteins. This gene product is thought to be involved in DNA double-strand break repair and DNA damage-induced checkpoint activation. [provided by RefSeq, Jul 2008]
- ReactivityHuman, Mouse
- Storage Instruction-20°C or -80°C,2°C to 8°C
- UNSPSC41116161

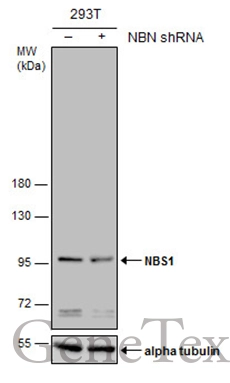
![Non-transfected (–) and transfected (+) 293T whole cell extracts (30 μg) were separated by 7.5% SDS-PAGE, and the membrane was blotted with NBS1 antibody [1C3] (GTX70222) diluted at 1:500. Non-transfected (–) and transfected (+) 293T whole cell extracts (30 μg) were separated by 7.5% SDS-PAGE, and the membrane was blotted with NBS1 antibody [1C3] (GTX70222) diluted at 1:500.](https://www.genetex.com/upload/website/prouct_img/normal/GTX70222/GTX70222_40004_20161103_WB_shRNA_watermark_w_23061221_703.webp)
![Various whole cell extracts (30 μg) were separated by 7.5% SDS-PAGE, and the membrane was blotted with NBS1 antibody [1C3] (GTX70222) diluted at 1:500. The HRP-conjugated anti-mouse IgG antibody (GTX213111-01) was used to detect the primary antibody. Various whole cell extracts (30 μg) were separated by 7.5% SDS-PAGE, and the membrane was blotted with NBS1 antibody [1C3] (GTX70222) diluted at 1:500. The HRP-conjugated anti-mouse IgG antibody (GTX213111-01) was used to detect the primary antibody.](https://www.genetex.com/upload/website/prouct_img/normal/GTX70222/GTX70222_45483_20240729_WB_25041720_566.webp)





![NBS1 antibody [1D7] detects NBS1 protein at nucleus by immunofluorescent analysis. Sample: A431 cells were fixed in 4% paraformaldehyde at RT for 15 min. Green: NBS1 stained by NBS1 antibody [1D7] (GTX70224) diluted at 1:500. Red: phalloidin, a cytoskeleton marker, diluted at 1:200.](https://www.genetex.com/upload/website/prouct_img/normal/GTX70224/GTX70224_40352_20200722_ICC_IF_w_23061221_571.webp)