Search results:
Product group Proteins / Signaling Molecules
Protein IDP22314
- SizePrice
Product group Proteins / Signaling Molecules
Protein IDP61077
- SizePrice
Product group Proteins / Signaling Molecules
UBE2L3 [UbcH7] (human) (rec.) (untagged)SBB-CE0020
Protein IDP68036
- SizePrice
Product group Proteins / Signaling Molecules
Protein IDP51668
- SizePrice
Product group Proteins / Signaling Molecules
Protein IDP61086
- SizePrice
Product group Proteins / Signaling Molecules
Protein IDP62837
- SizePrice
Product group Proteins / Signaling Molecules
Protein IDP22314
- SizePrice
Product group Proteins / Signaling Molecules
UcH-L3 (human) (rec.) (untagged)SBB-DE0023
Protein IDP15374
- SizePrice
Product group Proteins / Signaling Molecules
PLpro (SARS Coronavirus) (rec.) (His)SBB-DE0024
Protein IDP0C6U8
- SizePrice
Product group Proteins / Signaling Molecules
NEDP1 [SENP8] (human) (rec.) (His)SBB-DE0025
Protein IDQ96LD8
- SizePrice
Product group Proteins / Signaling Molecules
Protein IDQ9P0U3
- SizePrice
Product group Proteins / Signaling Molecules
20S Immunoproteasome (human) (untagged)SBB-PP0004
- SizePrice

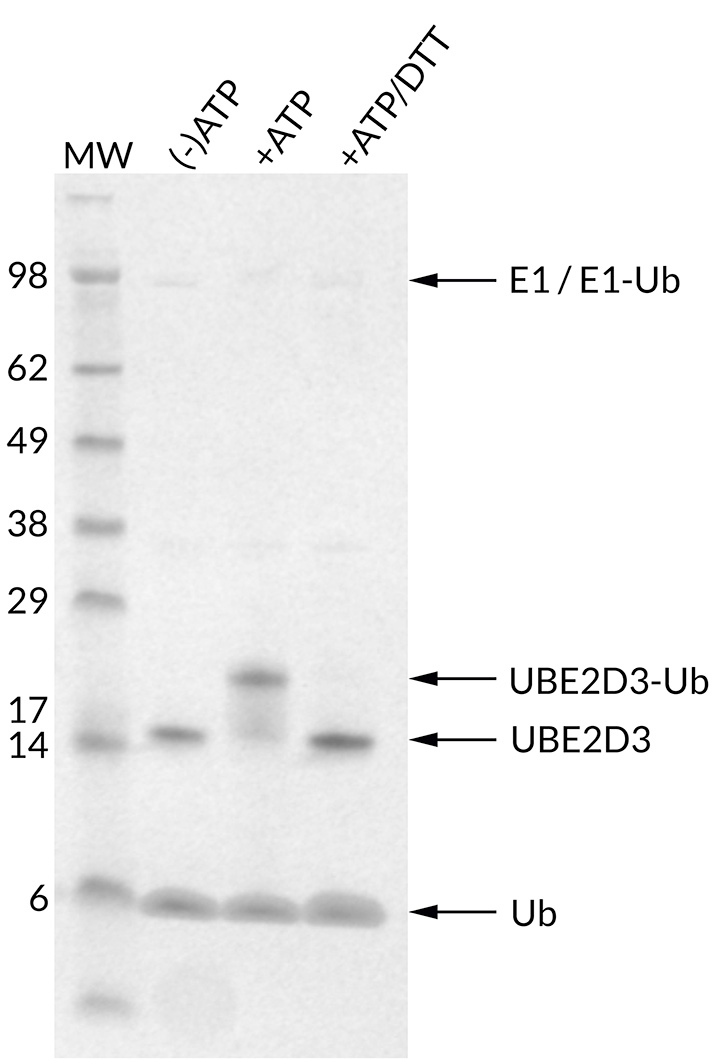
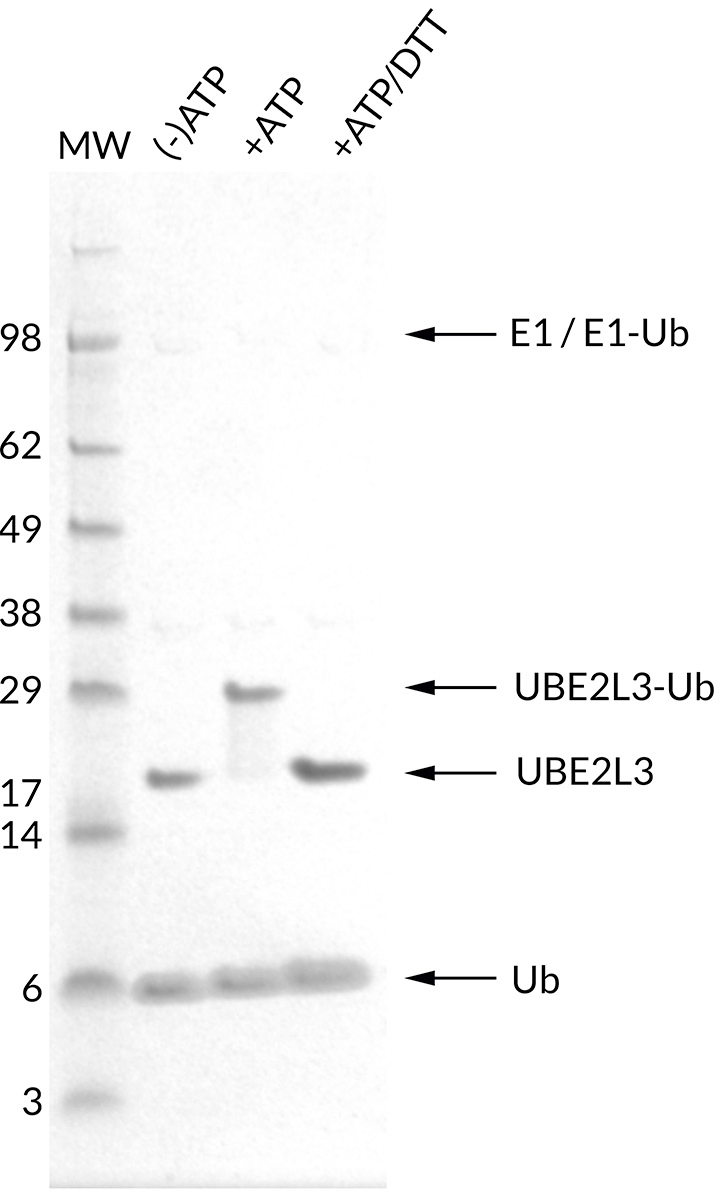
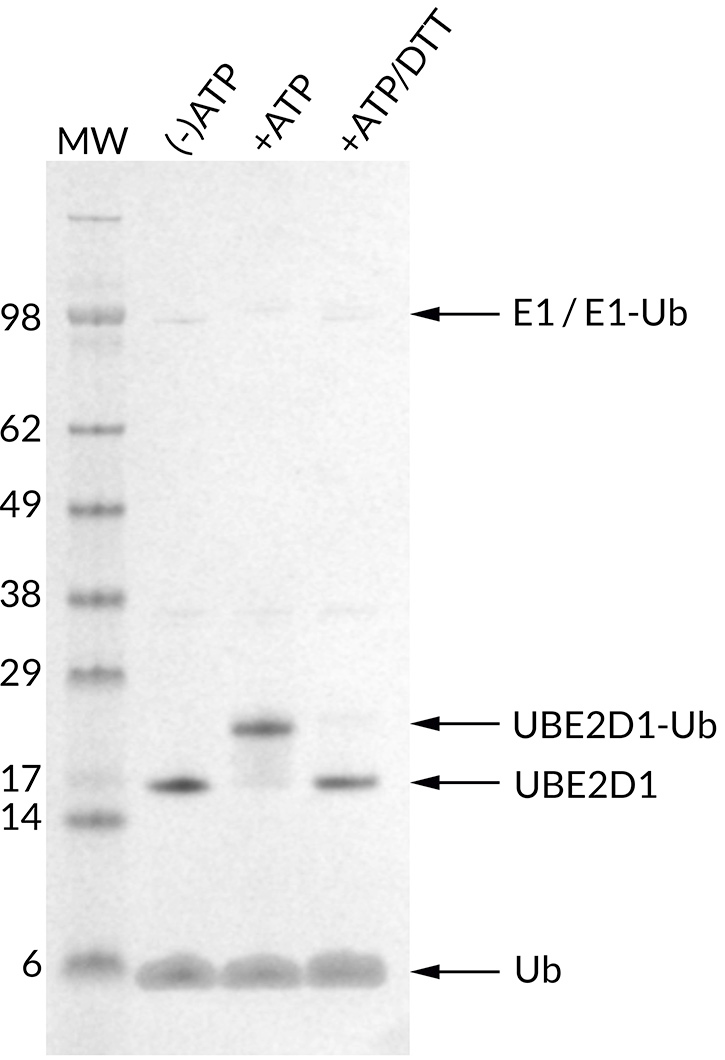
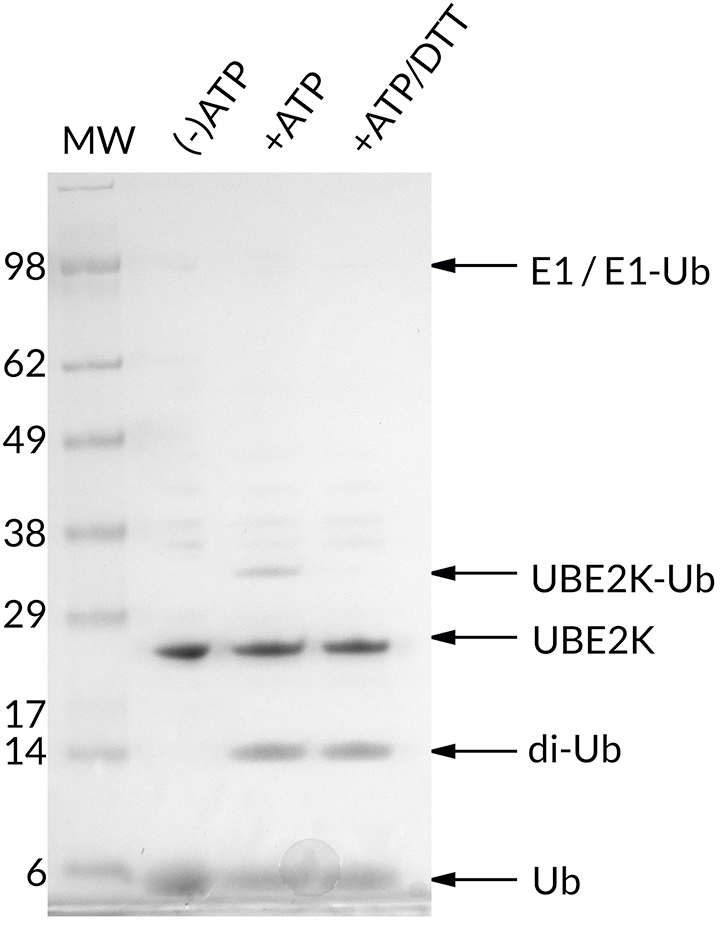
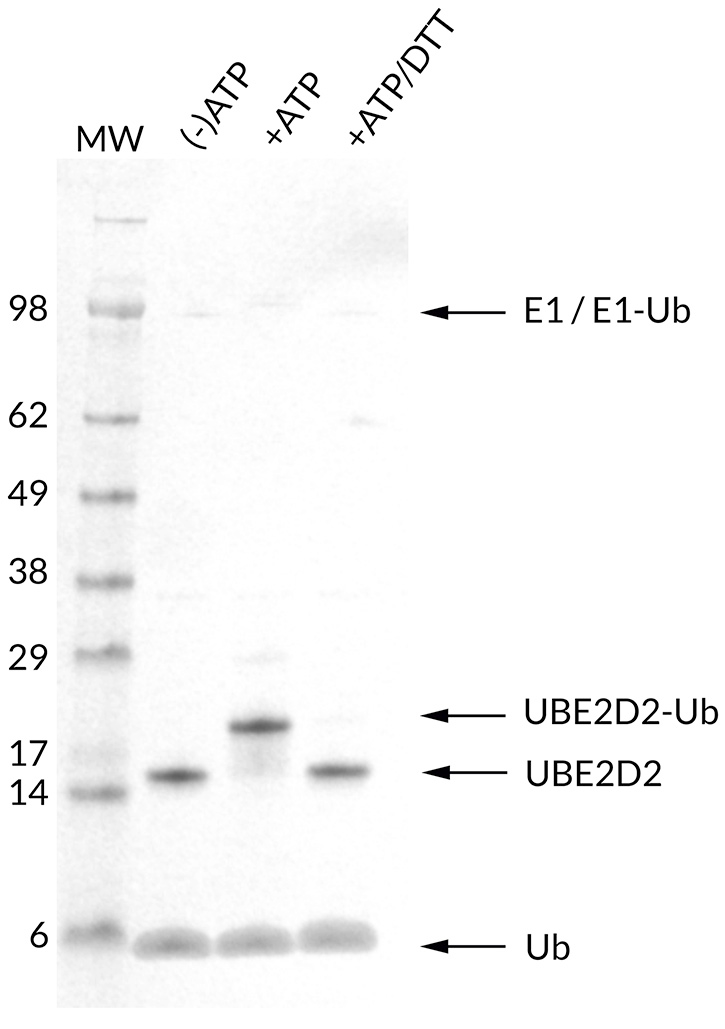
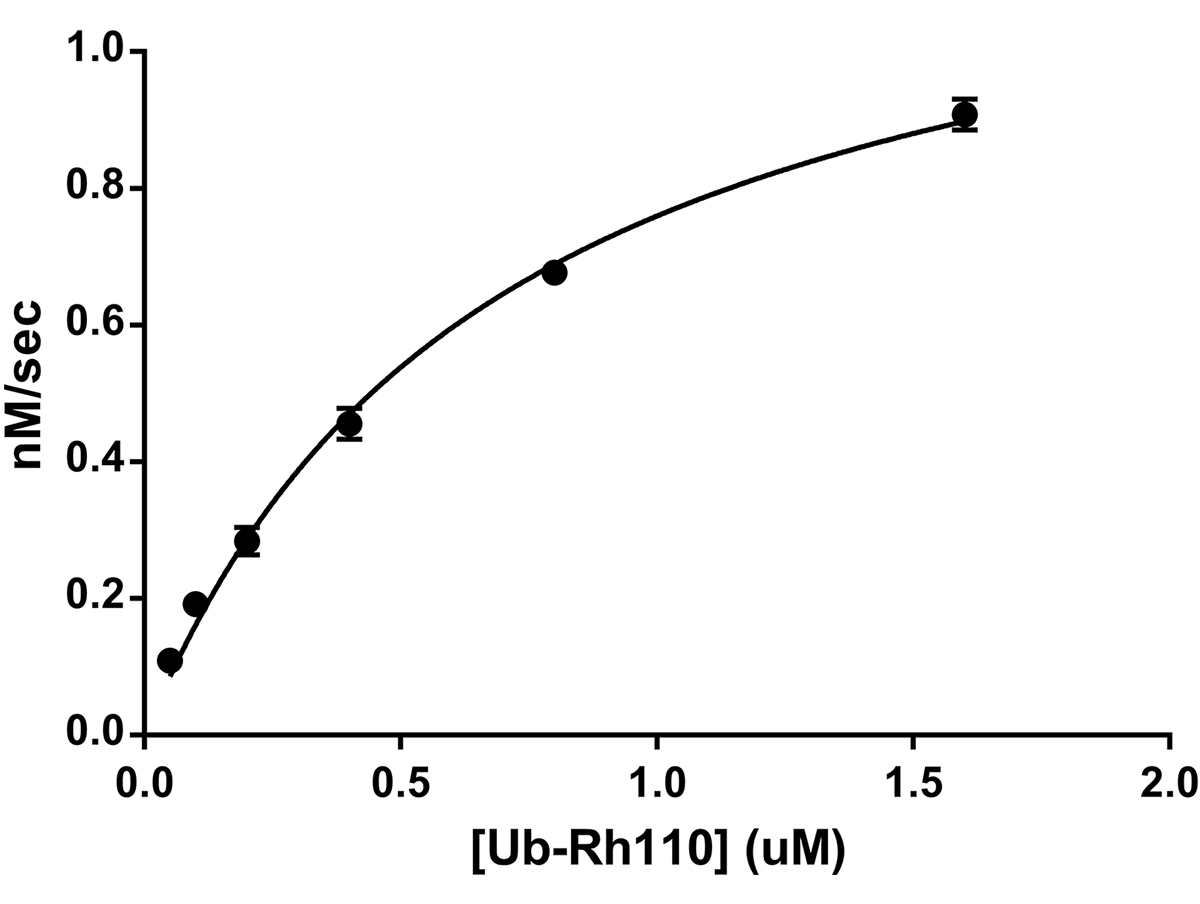
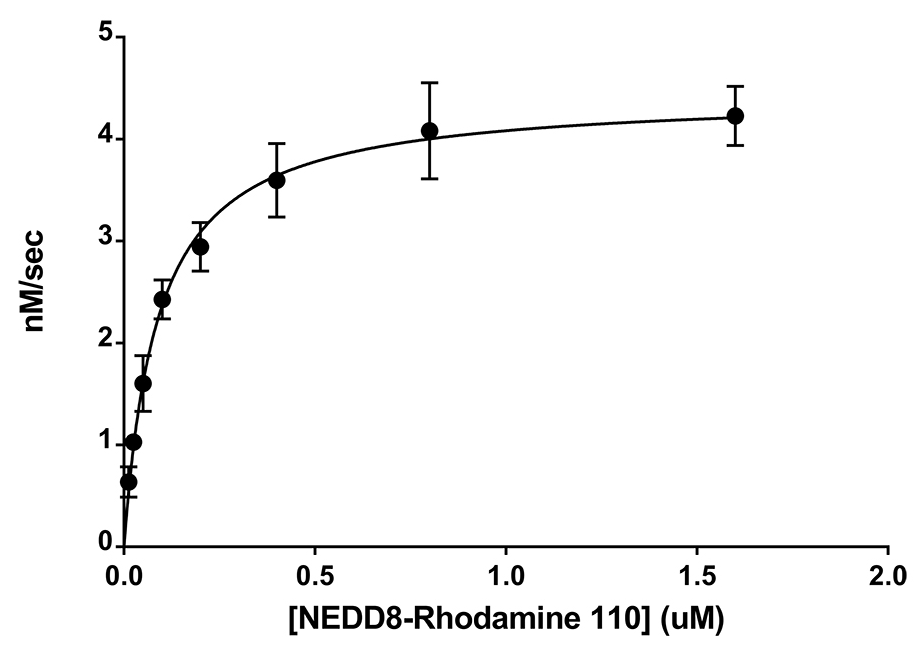
![Ubiquitin Activating Enzyme E1 [UBA1] (human) (rec.) (His)](https://adipogen.com/pub/media/catalog/product/s/b/sbb-ce0058_finalgel.png)